Starting from scratch: adaptation to variable environments after an extreme bottleneck
Project code: WE 2897/4-2

Inflorescences from the self-fertilizing Capsella rubella (left) and the outcrossing ancestor Capsella grandiflora (right)
While new species are usually formed through a gradual evolutionary process, they can also arise almost instantaneously, through events that involve severe genetic bottlenecking and loss of variability. Despite a very narrow genetic basis, such species can be remarkably successful. An important question is therefore how they manage to diversify and adapt to new habitats. We aim to address this problem by studying two very young plant species that underwent such bottlenecks: Arabidopsis suecica is in the same genus as the model plant Arabidopsis thaliana and originated as an interspecific hybrid between A. thaliana and A. arenosa. Capsella rubella, which belongs to a genus that is closely related to Arabidopsis, arose as a self-fertilizing form of an outcrossing ancestor. We use modern sequencing technologies to elucidate the patterns of variability in these species, both at the level of genomic DNA polymorphisms and in the transcribed portion of the genome, and compare it to the ancestral species. In addition, we perform functional and fitness studies. Taken together, our data will allow us to address important questions related to the evolutionary role of selfing and allopolyploidy.
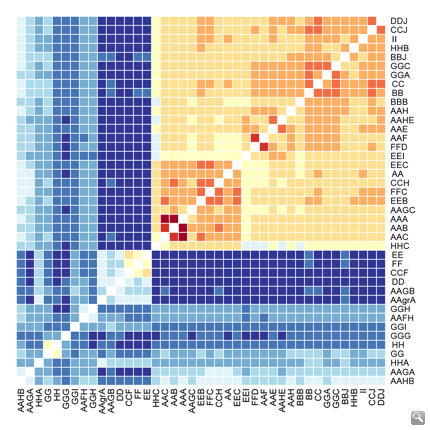
Whole genome resequencing of Capsella rubella individuals from throughout the range of the species identifies different populations based on overall genetic distance (lighter: more closely related; darker: more distantly related).
Publications related to the project
- Lee, C. R., Svardal, H., Farlow, A., Exposito-Alonso, M., Ding, W., Novikova, P., Alonso-Blanco, C., Weigel, D., Nordborg, M. (2017). On the post-glacial spread of human commensal Arabidopsis thaliana. Nat Commun 8:14458.


- The 1001 Genomes Consortium (2016). 1,135 genomes reveal the global of polymorphism in Arabidopsis thaliana. Cell 166(2):481-91.


- Grimm, D. G., Roqueiro, D., Salome, P., Kleeberger, S., Greshake, B., Zhu, W., Liu, C., Lippert, C., Stegle, O., Scholkopf, B., Weigel, D., Borgwardt, K. (2017). easyGWAS: A Cloud-based Platform for Comparing the Results of Genome-wide Association Studies. Plant Cell 29(1):5-19.


- Seren, Ü., Grimm, D. G., Fitz, J., Weigel, D., Nordborg, M., Borgwardt, K. M., Korte, A. (2016). A public database for Arabidopsis thaliana phenotypes. Nucl Acids Res 45(D1): D1054-D1059.


- Novikova, P. Y., J Hohmann, N., Nizhynska, V., Tsuchimatsu, T., Ali, J., Muir, G., Guggisberg, A., Paape, T., Schmid, K., Fedorenko, O. M., Holm, S., Sall, T., Schlotterer, C., Marhold, K., Widmer, A., Sese, J., Shimizu, K. K., Weigel, D., Kramer, U., Koch, M. A., Nordborg, M. (2016). Sequencing of the genus Arabidopsis identifies a complex history of nonbifurcating speciation and abundant trans-specific polymorphism. Nat Genet 48(9):1077-82.


- Kawakatsu, T., Huang, S. S., Jupe, F., Sasaki, E., Schmitz, R. J., Urich, M. A., Castanon, R., Nery, J. R., Barragan, C., He, Y., Chen, H., Dubin, M., Lee, C. R., Wang, C., Bemm, F., Becker, C., O'Neil, R., O'Malley, R. C., Quarless, D. X., Genomes Consortium, Schork, N. J., Weigel, D., Nordborg, M., Ecker, J. R. (2016). Epigenomic Diversity in a Global Collection of Arabidopsis thaliana Accessions. Cell 166(2):492-505.


- Weigel, D. and Nordborg, M. (2015). Population Genomics for Understanding in Wild Plant Species. Annu Rev Genet 49:315-38.


- Steige, K. A., Reimegard, J., Koenig, D., Scofield, D. G., Slotte, T. (2015). Cis-Regulatory Changes Associated with a Recent Mating System Shift and Floral Adaptation in Capsella. Mol Biol Evol 32(10):2501-14.


- Seymour, D. K., Koenig, D., Hagmann, J., Becker, C., Weigel, D. (2014). Evolution of DNA methylation patterns in the Brassicaceae is driven by differences in genome organization. PLoS Genet 10: e1004785.