Fakultät für
Chemie
und Biochemie
Biochemie II
Biomolekulare NMR-Spektroskopie
Prof. Dr. Raphael Stoll
Gebäude NC 5/171
Universitätsstraße 150
D-44780 Bochum
Tel.: +49 234 32-25466
Fax: +49 234 32-05466
E-Mail: bionmr@rub.de
Bioenergetics
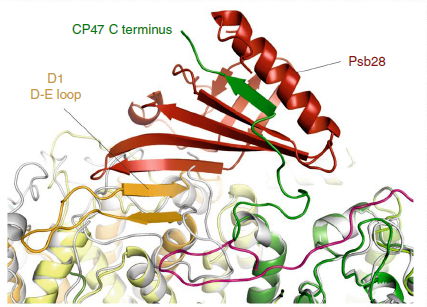
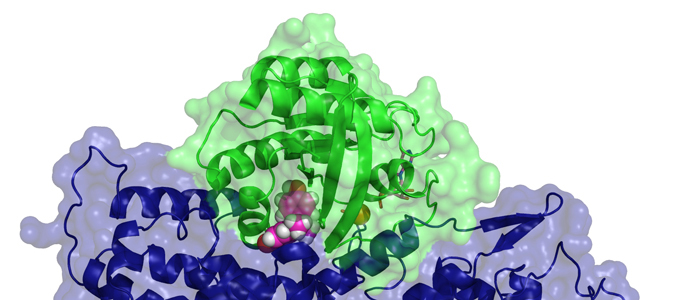
Biogenesis of photosystem II (PSII), nature's water-splitting catalyst, is assisted by auxiliary proteins that form transient complexes with PSII components to facilitate stepwise assembly events. Using cryo-electron microscopy, we solved the structure of such a PSII assembly intermediate from Thermosynechococcus elongatus at 2.94 Å resolution. It contains three assembly factors (Psb27, Psb28 and Psb34) and provides detailed insights into their molecular function. Binding of Psb28 induces large conformational changes at the PSII acceptor side, which distort the binding pocket of the mobile quinone (QB) and replace the bicarbonate ligand of non-haem iron with glutamate, a structural motif found in reaction centres of non-oxygenic photosynthetic bacteria. These results reveal mechanisms that protect PSII from damage during biogenesis until water splitting is activated. Our structure further demonstrates how the PSII active site is prepared for the incorporation of the Mn4CaO5 cluster, which performs the unique water-splitting reaction.
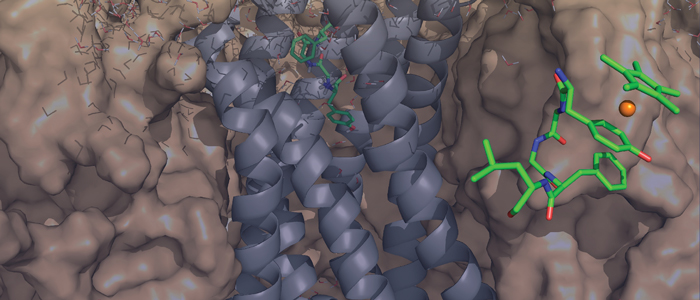
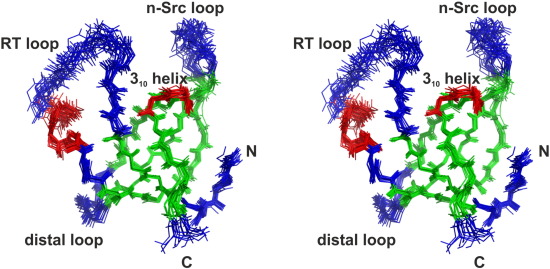 PetP is a peripheral subunit of the cytochrome b6f complex (b6f) present in both, cyanobacteria and red algae. It
is bound to the cytoplasmic surface of this membrane protein complex where it greatly affects the efficiency of
the linear photosynthetic electron flow although it is not directly involved in the electron transfer reactions.
Despite the crystal structures of the b6f core complex, structural information for the transient regulatory b6f subunits is still missing. Here we present the first structure of PetP at atomic resolution as determined by solution
NMR. The protein adopts an SH3 fold, which is a common protein motif in eukaryotes but comparatively rare
in prokaryotes. The structure of PetP enabled the identification of the potential interaction site for b6f binding
by conservation mapping. The interaction surface is mainly formed by two large loop regions and one short
310 helix which also exhibit an increased flexibility as indicated by heteronuclear steady-state {1H}-15N NOE
and randomcoil index parameters. The properties of this potential b6f binding site greatly differ fromthe canonical
peptide binding site which is highly conserved in eukaryotic SH3 domains. Interestingly, three other proteins
of the photosynthetic electron transport chain share this SH3 fold with PetP: NdhS of the photosynthetic NADH
dehydrogenase-like complex (NDH-1), PsaE of the photosystem 1 and subunit α of the ferredoxin–thioredoxin
reductase have, similar to PetP, a great impact on the photosynthetic electron transport. Finally, a model is presented to illustrate how SH3 domains modulate the photosynthetic electron transport processes in
cyanobacteria.
PetP is a peripheral subunit of the cytochrome b6f complex (b6f) present in both, cyanobacteria and red algae. It
is bound to the cytoplasmic surface of this membrane protein complex where it greatly affects the efficiency of
the linear photosynthetic electron flow although it is not directly involved in the electron transfer reactions.
Despite the crystal structures of the b6f core complex, structural information for the transient regulatory b6f subunits is still missing. Here we present the first structure of PetP at atomic resolution as determined by solution
NMR. The protein adopts an SH3 fold, which is a common protein motif in eukaryotes but comparatively rare
in prokaryotes. The structure of PetP enabled the identification of the potential interaction site for b6f binding
by conservation mapping. The interaction surface is mainly formed by two large loop regions and one short
310 helix which also exhibit an increased flexibility as indicated by heteronuclear steady-state {1H}-15N NOE
and randomcoil index parameters. The properties of this potential b6f binding site greatly differ fromthe canonical
peptide binding site which is highly conserved in eukaryotic SH3 domains. Interestingly, three other proteins
of the photosynthetic electron transport chain share this SH3 fold with PetP: NdhS of the photosynthetic NADH
dehydrogenase-like complex (NDH-1), PsaE of the photosystem 1 and subunit α of the ferredoxin–thioredoxin
reductase have, similar to PetP, a great impact on the photosynthetic electron transport. Finally, a model is presented to illustrate how SH3 domains modulate the photosynthetic electron transport processes in
cyanobacteria.
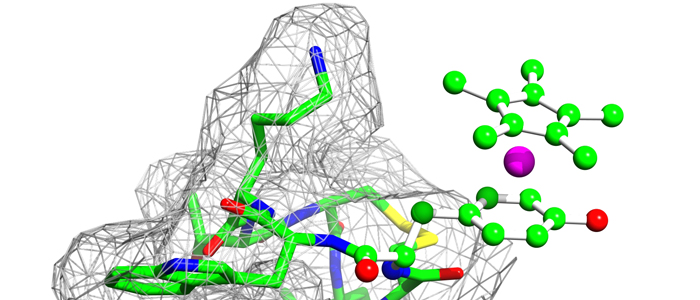
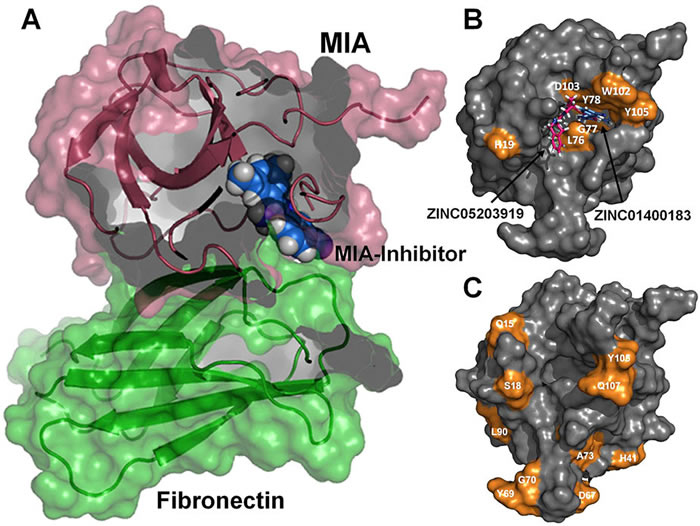
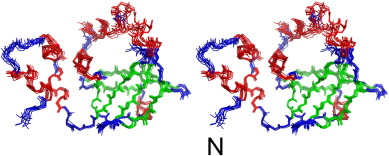 The cyanobacterial multi-subunit membrane protein complex NDH-1 is both structurally and functionally related to Complex I of eubacteria and mitochondria. In addition to functions in respiration and cyclic electron transfer around photosystem I (PSI), the cyanobacterial NDH-1 complex is involved in a unique mechanism for inorganic carbon concentration. Although the crystal structures of the similar respiratory Complex I from Thermus thermophilus and Escherichia coli are known, atomic structural information is not available for the cyanobacterial NDH-1 complex yet. In particular, the structures of those subunits that are not homologous to Complex I will help to understand their distinct functions. The 15.7kDa protein CupS is a small soluble subunit of the complex variant NDH-1MS, which is thought to play a role in CO2 conversion. Here, we present the NMR structure of CupS from Thermosynechococcus elongatus, which is the very first structure of a specific cyanobacterial NDH-1 complex subunit. CupS shares a structural similarity with members of the Fasciclin protein superfamily. The structural comparison to Fasciclin type proteins based on known NMR structures and protein sequences of human TGFBIp, MPB70 from Mycobacterium bovis, and Fdp from Rhodobacter sphaeroides, together with a virtual docking model of CupS and NdhF3, provide first insight into the specific binding of CupS to the NDH-1MS complex at atomic resolution.
The cyanobacterial multi-subunit membrane protein complex NDH-1 is both structurally and functionally related to Complex I of eubacteria and mitochondria. In addition to functions in respiration and cyclic electron transfer around photosystem I (PSI), the cyanobacterial NDH-1 complex is involved in a unique mechanism for inorganic carbon concentration. Although the crystal structures of the similar respiratory Complex I from Thermus thermophilus and Escherichia coli are known, atomic structural information is not available for the cyanobacterial NDH-1 complex yet. In particular, the structures of those subunits that are not homologous to Complex I will help to understand their distinct functions. The 15.7kDa protein CupS is a small soluble subunit of the complex variant NDH-1MS, which is thought to play a role in CO2 conversion. Here, we present the NMR structure of CupS from Thermosynechococcus elongatus, which is the very first structure of a specific cyanobacterial NDH-1 complex subunit. CupS shares a structural similarity with members of the Fasciclin protein superfamily. The structural comparison to Fasciclin type proteins based on known NMR structures and protein sequences of human TGFBIp, MPB70 from Mycobacterium bovis, and Fdp from Rhodobacter sphaeroides, together with a virtual docking model of CupS and NdhF3, provide first insight into the specific binding of CupS to the NDH-1MS complex at atomic resolution.
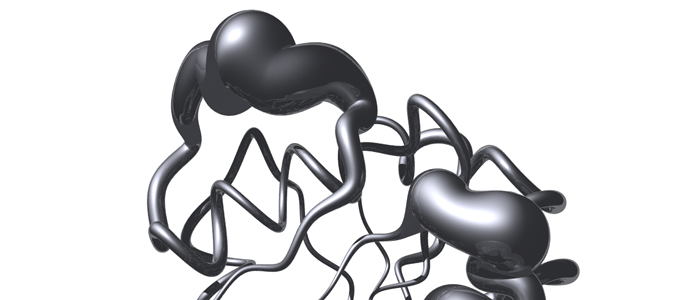
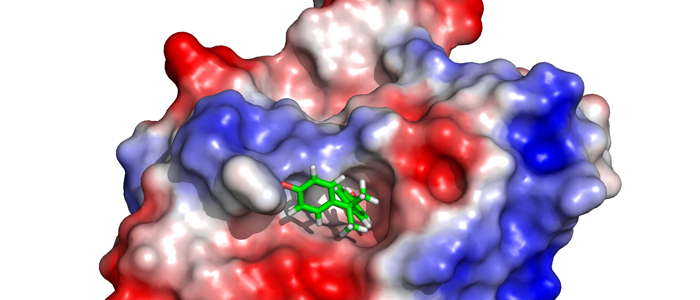
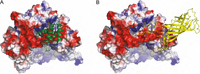 Psb27 is a membrane-extrinsic subunit of photosystem II (PSII) where it is involved in the assembly and maintenance of this large membrane protein complex that catalyzes one of the key reactions in the biosphere, the light-induced oxidation of water. Here, we report for the first time the structure of Psb27 that was not observed in the previous crystal structures of PSII due to its transient binding mode. The Psb27 structure shows that the core of the protein is a right-handed four-helix bundle with an up-down-up-down topology. The electrostatic potential of the surface generated by the amphipathic helices shows a dipolar distribution which fits perfectly to the major PsbO binding site on the PSII complex. Moreover, the presented docking model could explain the function of Psb27 (A), which prevents the binding of PsbO to facilitate the assembly of the Mn4Ca cluster (B).
Psb27 is a membrane-extrinsic subunit of photosystem II (PSII) where it is involved in the assembly and maintenance of this large membrane protein complex that catalyzes one of the key reactions in the biosphere, the light-induced oxidation of water. Here, we report for the first time the structure of Psb27 that was not observed in the previous crystal structures of PSII due to its transient binding mode. The Psb27 structure shows that the core of the protein is a right-handed four-helix bundle with an up-down-up-down topology. The electrostatic potential of the surface generated by the amphipathic helices shows a dipolar distribution which fits perfectly to the major PsbO binding site on the PSII complex. Moreover, the presented docking model could explain the function of Psb27 (A), which prevents the binding of PsbO to facilitate the assembly of the Mn4Ca cluster (B).