DNA Block Copolymers as Programmable Containments in Microfluidic Devices
PI: Prof. Andreas Herrmann, University of Groningen, The Netherlands, a.herrmann@rug.nl
The main objective of workpackage 2 within the ECCell project is to construct containments that can be employed and manipulated within microfluidic devices. The central chemical building block with which this goal is realized is DNA block copolymers.
Amphiphilic block copolymers consist of alternating hydrophobic and hydrophilic polymer segments. These materials tend to undergo microphase separation. Depending on the block length and block length ratios different nanostructures are adopted. In bulk the morphologies range from spherical, through cylindrical and gyroidal, to lamellar structures. In solution typically micelles or vesicular assemblies are formed. The use of block copolymers as supramolecular building blocks allows to adjust the dimensions and the stability of the self-assembled objects very precisely due to several adjustable structural parameters of the constituent components. Recently a novel class of amphiphilic block copolymer systems was introduced that consists of bioorganic hybrids.[1] Synthetic hydrophobic polymer blocks were combined with nucleic acid units in a covalent fashion to form DNA block copolymers (DBCs). Due to the presence of the oligonucleotide segment these structures are programmable supramolecular building blocks. By hybridization spherical DBC aggregates were transformed into rod-like micelles.[2] Moreover, DBC micelles were employed as antisense reagents and nanoscopic scaffolds for various DNA-templated organic reactions.[3] They could even be utilized for targeted drug delivery.[4] The DBC nanocontainers were loaded with a hydrophobic anti-cancer drug that accumulated in the core of the micelles. Additionally, the nucleic acid corona of the micelles was equipped with targetting moieties that recognize cancer cells by a simple hybridization procedure. When both, the targeting units and the drug, acted together within the DBC micelle scaffold cancer cells could be efficiently killed.
Here three types of containments are realized within the ECCell project that go beyond simple micelle assemblies as they were described above, i.e. DBC vesicles, DBC micelle networks and DBC-templated formation of virus capsids.
DBC vesicles
Two types of vesicles were realized within the ECCell project. In a first approach, a diblock copolymer that was known to adopt vesicle structures was functionalized with an oligonucleotide by our well established grafting onto approach on the solid phase. The resulting triblock structures, polyisorene-block-polyethyleneoxide-block-DNA, adopted vesicles with sizes in the low micrometer range. The formation of the structures was proven by light and fluorescence microscopy by selective staining of the nucleic acid segments within the aggregates (Figure 1A). In a second approach amphiphilic DBCs were incorporated into giant unilammelar vesicles the membrane of which was composed of phospholipids. By DNA hybridization vesicles encoded with complementary sequences were transformed into vesicle aggregates. Thereby, multiple hybridization events between two vesicles lead to hemifusion resulting in the formation of planar arrangements of the vesicle membranes (Figure 1B).
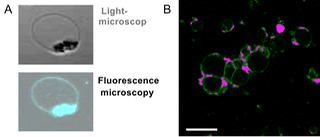
Figure 1: Vesicle architectures containing DBCs. A: Vesicles prepared exclusively from DNA triblock copolymers. B: Vesicle aggregation induced by DBCs incorporated into the vesicle membrane. (scale bar: 25 μm)
DBC micelle networks
Another way of immobilizing and compartmentalizing materials within microfluidic channels is the reversible formation of DBC micelle networks. For that purpose amphiphilic DBCs were fabricated and transformed into micelles. By the addition of complementary DNA that hybridizes to DNA nanoparticles with different sequence compositions hydrogels originate that are cross-linked by two non-covalent structural motifs, i.e. hydrophobic interactions of the polymers and Watson-Crick base pairing (Figure 2). The crossl-inking DNA can be reversibly released with the help of removal strands (not shown).
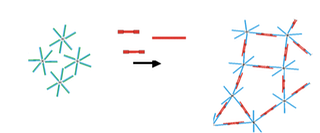
Figure 2: Schematic representation of the formation of a DBC micelle network.
DBCs within virus capsids
Virus Capsids are fascinating multifunctional containers evolved by nature to effectively deliver nucleic acids inside cells. Throughout the ECCell project we developed a general loading strategy of these biological containments. This is especially important since virus capsids have recently gained great interest with potential drug delivery and gene therapy applications. Cowpea Chlorotic Mottle Virus Capsids were found to efficiently envelope DBC micelles. Without the charged DBC templates the capsid proteins exists as dimers and do not form a closed shell. Due to the presence of the DBC micelles inside the viral containments hydrophilic compounds can be co-internalized by chemical attachment to complementary oligonucleotides and hybridization. Hydrophobic moieties are loaded into the hydrophobic core of the DBC micelles prior to templating the capsid formation (Figure 3).
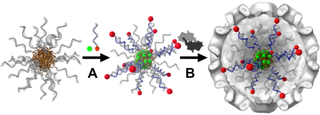
Figure 3: Schematic representation of a general loading strategy of virus capsids. A: DBC micelles are loaded either with a hydrophilic moiety (red) or a hydrophobic entity (green). B: Incorporation of these compounds into the capsids is achieved by incubating with protein dimers.
[1] F. E. Alemdaroglu, A. Herrmann, Organic & Biomolecular Chemistry 2007, 5, 1311.
[2] K. Ding, F. E. Alemdaroglu, M. Boersch, R. Berger, A. Herrmann, Angew. Chem. Int. Ed. 2007, 46, 1172.
[3] F. E. Alemdaroglu, K. Ding, R. Berger, A. Herrmann, Angew. Chem. Int. Ed. 2006, 45, 4206.
[4] F. E. Alemdaroglu, C. N. Alemdaroglu, P. Langguth, A. Herrmann, Adv. Mat. 2008, 20, 899.
