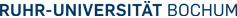Genetic tumor progression model for PDAC
Luttges J, Galehdari H, Brocker V, Schwarte-Waldhoff I, Henne-Bruns D, Kloppel G, Schmiegel W, Hahn SA. ![]()
Allelic loss is often the first hit in the biallelic inactivation of the p53 and DPC4 genes during pancreatic carcinogenesis. Am J Pathol. 2001 May;158(5):1677-83. PMID: 11337365
In a collaborative study with Prof. Lüttges and Prof. Klöppel, Institute of Pathology, Kiel supported by the German Cancer Foundation (Deutsche Krebshilfe) we tested whether molecular genetic alterations can be correlated with the histopathological PanIN classification and may help to further categorize the various PanIN grades. We determined the frequencies of allelic loss at chromosomal arms 9p, 17p, and 18q in 81 microdissected duct lesions of various PanIN grades, using a combination of whole genome amplification and microsatellite analysis. In addition we examined the p53 and Dpc4 protein expression patterns by immunohistochemical analysis. In PanIN-1, we did not detect allelic losses. In PanIN-2, allelic losses were found in increasing frequency, and were particularly high in those lesions with moderate-grade dysplasia (low grade, 20, 33, and 17%, loss at 9p, 17p, and 18q, respectively; moderate grade, 46, 77, and 58%). PanIN-3 and invasive carcinomas exhibited abundant losses. Abnormal p53 and Dpc4 protein expression was only rarely identified in PanIN-2 lesions, but occurred frequently in PanIN-3 lesions and invasive carcinomas. Thus, we found a progressive increase in allelic losses at all three chromosomal regions in PanINs from low to high grade dysplasia, with the highest rate of allelic loss identified in carcinomas (Figure 1A). The majority (75%) of PanIN-3 lesions revealed losses at two or three of the chromosomal regions tested, whereas 67% of PanIN-2 lesions with moderate dysplasia had losses at one or two of the three regions (Figure 1B). These data are in agreement with earlier molecular studies showing that the successive accumulation of genetic changes paralleled the severity of ductal dysplasia. Our data also confirm the previously suggested precursor nature of the ductal lesions and the proposed tumor progression model for pancreatic neoplasia. Furthermore, the combined genetic and protein expression data support a model in which allelic loss is the first hit in the biallelic inactivation of the p53 and DPC4 tumor suppressor genes. In addition, our data indicate that allelic loss analysis may be useful in separating PanIN-2 lesions with low-grade dysplasia from those PanIN-2 lesions with moderate-grade dysplasia, each potentially representing a distinct progression step toward invasive carcinoma. The lack of LOH in PanIN-1 lesions precludes within this study the genetic proof that PanIN-1 lesions are an early step in pancreatic carcinoma progression.
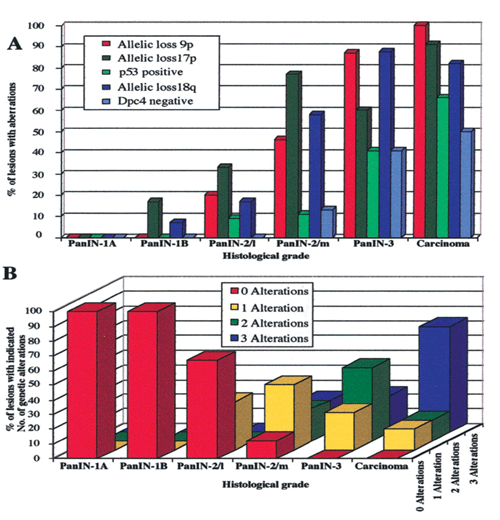
Figure 1: Molecular analyses of various grades of PanIN.
A: Frequencies of allelic loss at chromosomal regions 9p, 17p, and 18q together with frequencies of nuclear p53 expression and lack of Dpc4 protein expression. B: Accumulation of genetic alterations. The number of alterations refers to the number of allelic losses at three chromosomal regions (9p, 17p, and 18q) in those lesions that were informative for at least one marker at each of the three loci. PanIN-2/L, PanIN-2 lesion with low-grade dysplasia; PanIN-2/m, PanIN-2 lesion with moderate-grade dysplasia.
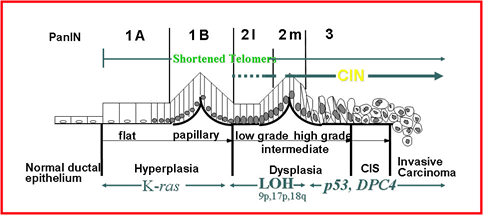
Figure 2: Schematic model of the pancreatic tumor progression.